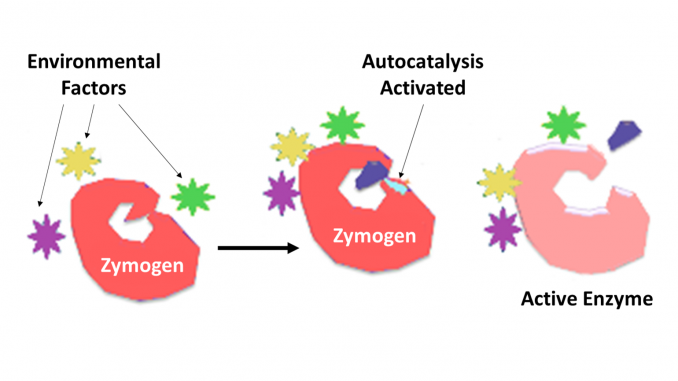
Zymogen, also called proenzyme, any of a group of proteins that display no catalytic activity but are transformed within an organism into enzymes, especially those that catalyze reactions involving the breakdown of proteins. The biochemical change usually occurs in Golgi bodies, where a specific part of the precursor enzyme is cleaved in order to activate it. The inactivating piece which is cleaved off can be a peptide unit, or can be independently folding domains comprising more than 100 residues. Although they limit the enzyme’s ability, these n-terminal extensions of the enzyme or a “prosegment” often aid in the stabilizing and folding of the enzyme they inhibit.
It is now well recognized that proteolytic enzymes play key roles in the regulation of or control over the action of other proteins. Such enzymes can be found in all species from bacteria to man and in control of diverse systems, which include hormone production, bacteriophage assembly, development, fertilization, digestion, defense against invading organisms, and tissue repair. In most cases the proteolytic enzymes are known to be synthesized as inactive precursor proenzymes, or zymogens. They are activated by proteolytic cleavage of a single peptide bond in the proenzyme and so become catalytically active. Further control over the degree of specificity for a target molecule or molecules is determined by the degree of specificity inherent to the enzyme. Further control over the time and location of action is often carried out by protein inhibitors of the requisite specificity.
The structure of trypsinogen is generally much closer to that of trypsin than is chymotrypsinogen to chymotrypsin, The structure of trypsinogen does not exclude the possibility of substrate binding in a mode similar to that found for trypsin, although changes in the structure of this region contribute to an impaired or altered substrate binding mode-certainly for benzamidine and most probably for a substrate side chain, If the proenzyme is considered to be rigid, then the general base catalyst and the oxyanion binding site formed by the N-H groups of Gly193 and Ser195 are too far apart to cooperate in substrate hydrolysis. Even if the proenzyme structure were to change on substrate binding, as seems likely, nonproductive binding may provide another important component for zymogen inactivity. Such binding might still leave too great a distance, or an unfavorable interaction, between the substrate and elements of the catalytic center. The altered position of the chain between Lys188A and Ser195 and the main chain between Trp215 and Ser217 could be responsible for competitive yet nonproductive substrate binding, as could the N-H group of Gly193 since they are normally involved in orienting the substrate. These possibilities rematin as prime candidates in a universal scheme for inactivity of the trypsinogen-like zymogens.
The pancreas secretes zymogens partly to prevent the enzymes from digesting proteins in the cells in which they are synthesized. Enzymes like pepsin are created in the form of pepsinogen, an inactive zymogen. Pepsinogen is activated when chief cells release it into the gastric acid, whose hydrochloric acid partially activates it. Another partially activated pepsinogen completes the activation by removing the peptide, turning the pepsinogen into pepsin. Accidental activation of zymogens can happen when the secretion duct in the pancreas is blocked by a gallstone resulting in acute pancreatitis.
Fungi also secrete digestive enzymes into the environment as zymogens. The external environment has a different pH than inside the fungal cell and this changes the zymogen’s structure into an active enzyme.
Another way that enzymes can exist in inactive forms and later be converted to active forms is by activating only when a cofactor, called a coenzyme, is bound. In this system, the inactive form (the apoenzyme) becomes the active form (the holoenzyme) when the coenzyme binds.
Reference:
Stroud, R.M, Kossiakoff, A.A, Chambers, J.L. Mechanisms of zymogen activation. Annual review of biophysics and bioengineering, 1977, 6(1): 177-193.
Related Products at Creative Enzymes:

Leave a Reply